3D Probabilistic Modeling of Human Stem Cell Organization
As part of the Allen Integrated Cell, we have developed and implemented a state-of-the-art machine learning model, the 3D Probabilistic Cell Model, which captures the relative variations in cell and organelle morphologies and locations for all components studied. Like traditional probabilistic approaches, this model allows us to analyze heterogeneity in our cell population – with a powerful difference.
The model can capture and analyze all of the variation among components of our cells and then use this information to predict the locations of structures not observed in any particular sample, given the location and morphology of the cell boundary and the nucleus. In addition, the model allows us to both predict how cells and their components will look given certain conditions, and to integrate cells with components observed in different measurements (see preprint Building a 3D Integrated Cell, December 2017).
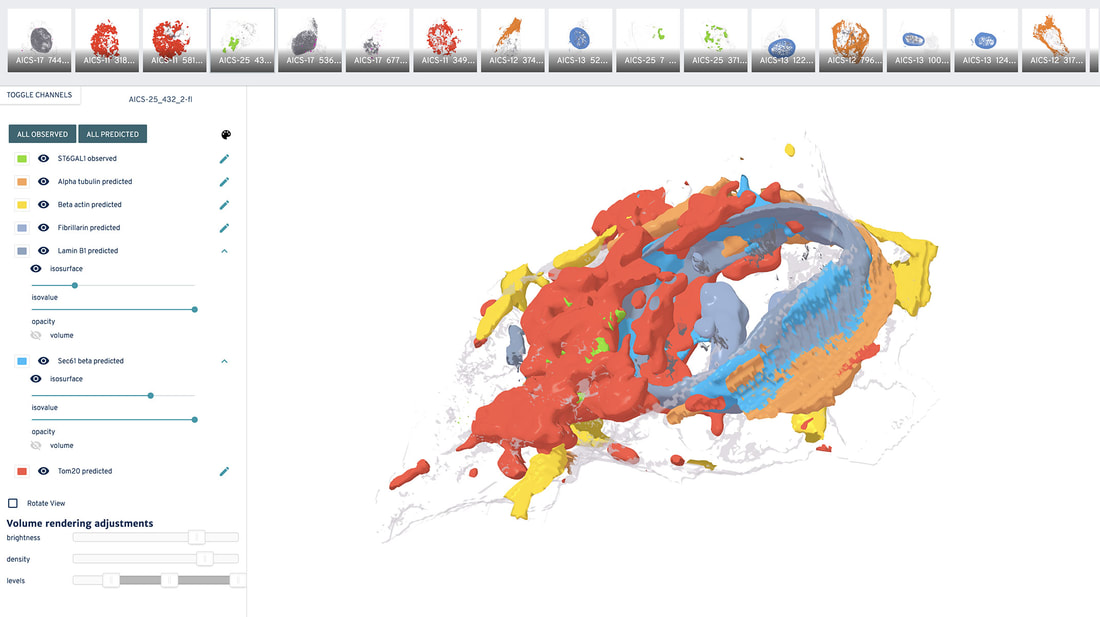
Explore how organelles are likely shaped and where these are most probably located, as well as probabilistic spatial distributions of these structures in the 3D viewer below:
Explore how organelles are likely shaped and where these are most probably located, as well as probabilistic spatial distributions of these structures in the 3D viewer below:
How does the 3D probabilistic cell structure model work?
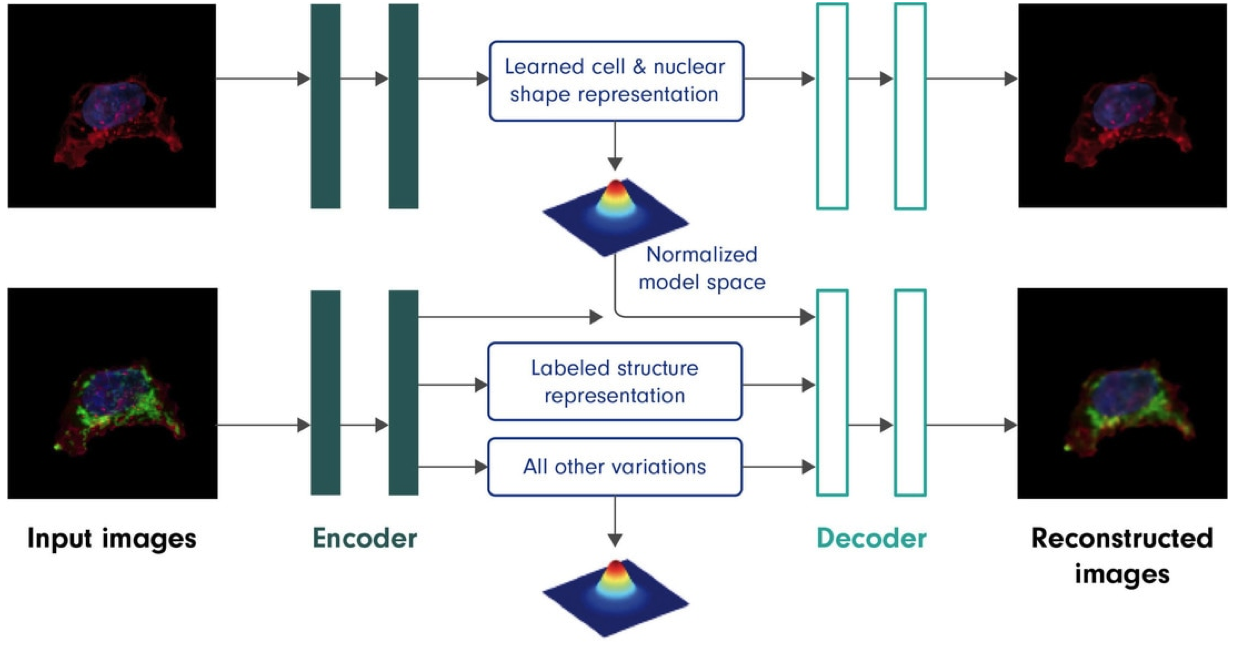
Recent machine learning methods based on deep neural networks (deep learning) are a powerful approach for encoding and integrating large sets of diverse images, and then generating integrated photorealistic outputs. Here, we have developed and applied a novel computational method (see Figure), to predict the location and morphologies of key organelles in our cells, given an observed cell (plasma membrane) and nuclear (DNA) morphology.
Figure. Autoencoders can learn conditional models. Overview of the deep learning model used to encode cell variance and predict key proteins in a new cell. The model captures the variance between cell and nucleus (DNA) location and shape, and also serves to capture covariance between these (cell and nucleus) as well as observed structures, e.g., nuclear membrane (LaminB1), endoplasmic reticulum (Sec61b), mitochondria (Tom20), microtubules (alpha-tubulin), actin (alpha-actinin; beta-actin), tight junctions (ZO-1), (arXiv:1511.05644v2 [cs.LG]. The model latent spaces, as encapsulated in the blue text boxes above, encode, from top, a learned cell and nuclear shape representation, a structure label (class, e.g. mitochondria (Tom20)), and all other variation in the structure shape and location, e.g. possible morphology and location of mitochondria (Tom20).
It seems like the predicted channels from the probabilistic model do not always match up with the observed channels – why is that?
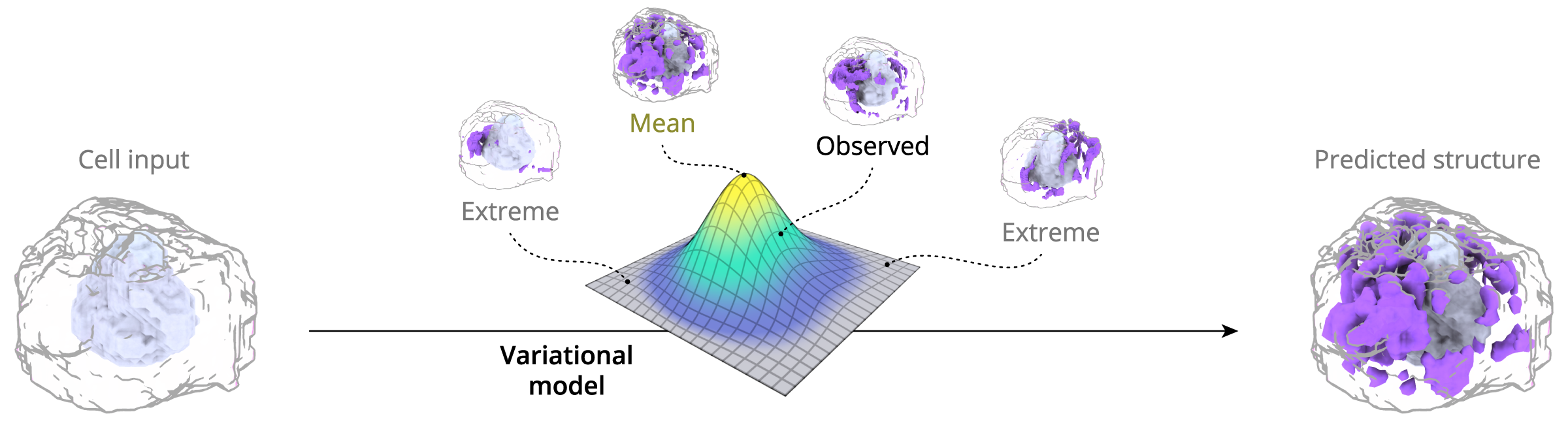
The probabilistic model provides an organelle’s most likely location and shape in any given cell, based on what has been observed for all cells. Therefore, the specific prediction will likely not match up with any particular single instance of measured data. This is expected behavior for models of this type.
Other resources
|
Links to our repositories, where all our open-source tools and code can be found.
|
|
The latest publications & preprints.
|
|
Detailed and technical methods from the publications provided here
|