Overview of the Allen Institute for Cell Science workflow
The mission of the Institute is to understand and predict cell behaviors. The current project aims to:
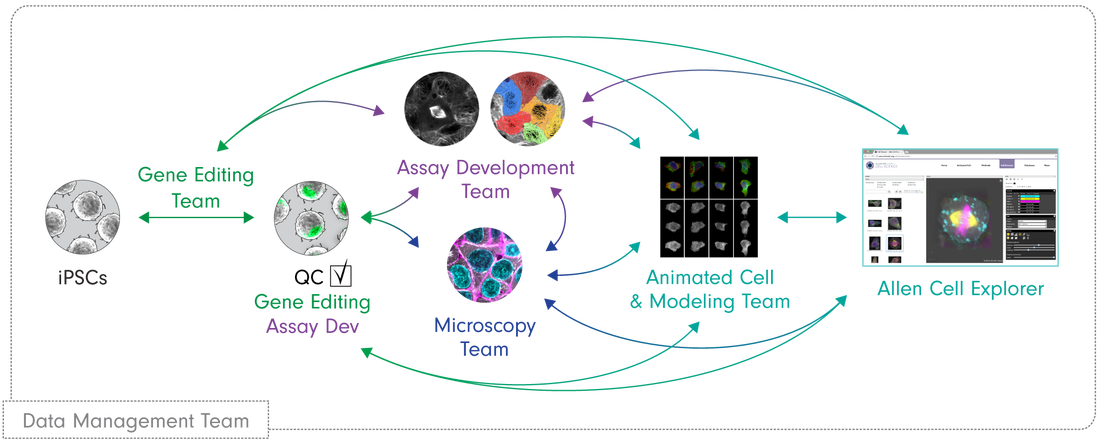
We have implemented this ambitious project through an integrated workflow employing interdisciplinary teams all working toward a common goal. This section outlines the elements of this highly interactive and integrated workflow, shown in the figure below.
- create a stem cell state space by conjoining genomic, imaging, and phenotypic data from cells that are differentiating, that are at different points in the cell cycle, or that have been perturbed
- determine the mechanisms by which cells transition from state to state
- determine how the expression of a subset of genes produces key cellular states.
We have implemented this ambitious project through an integrated workflow employing interdisciplinary teams all working toward a common goal. This section outlines the elements of this highly interactive and integrated workflow, shown in the figure below.
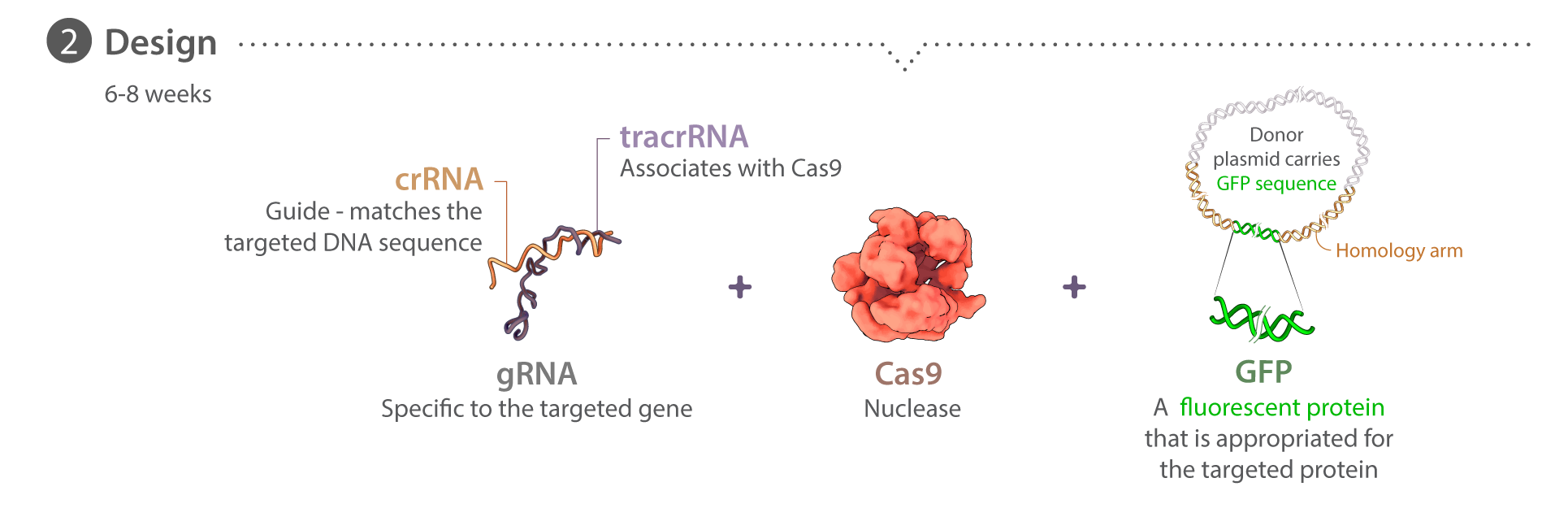
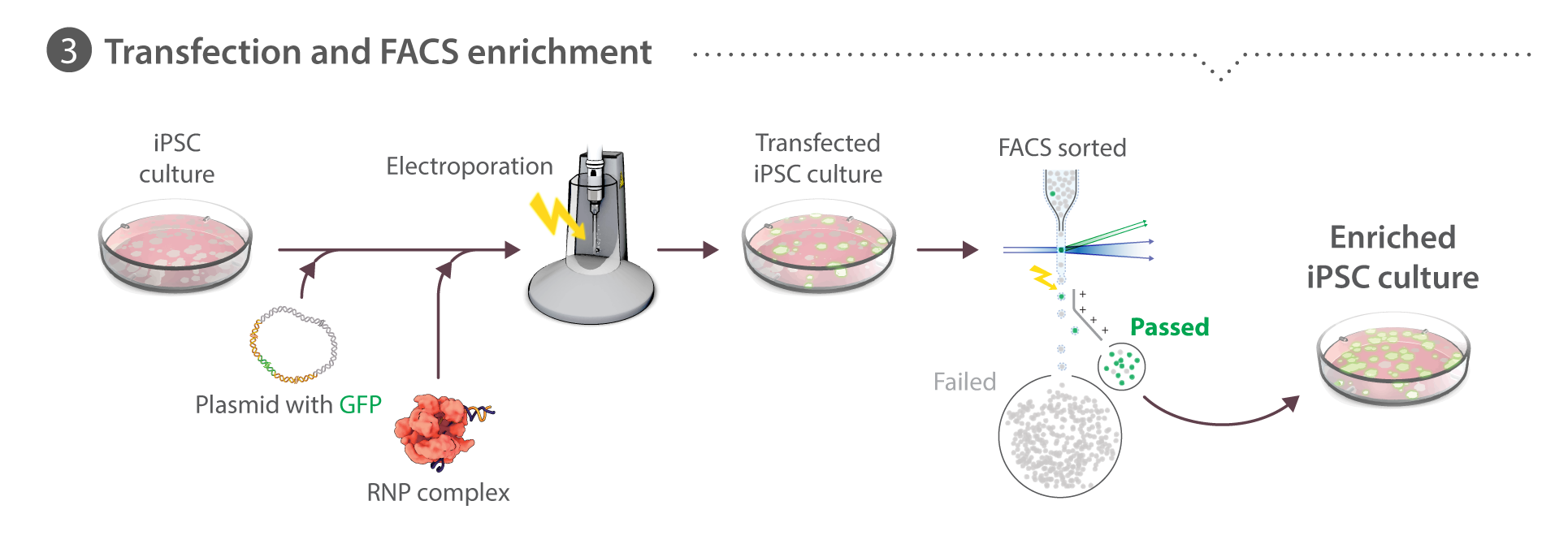
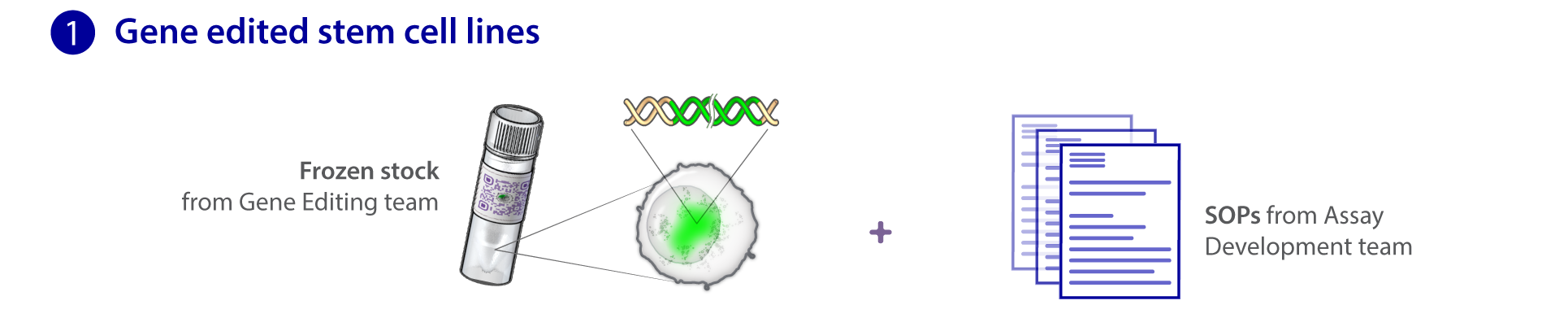
Our pipeline starts with the Gene Editing team, which creates unique cell lines with certain structures tagged with a fluorescent molecule. With a joint effort from the Gene Editing and Assay Development teams, these cell lines undergo a rigorous process of quality control and testing to ensure they behave and develop normally. The Microscopy team takes images and videos of the cells. Together with the Assay Dev team, they develop tools to analyze these media data to make sense of the cells' structures, behaviors and organization. Subsequently, the Modeling team uses the generated data to build models to learn and predict the same properties for future simulation purposes. All of our methods and data are available to the public here at the Allen Cell Explorer.
Stem cells & gene editing
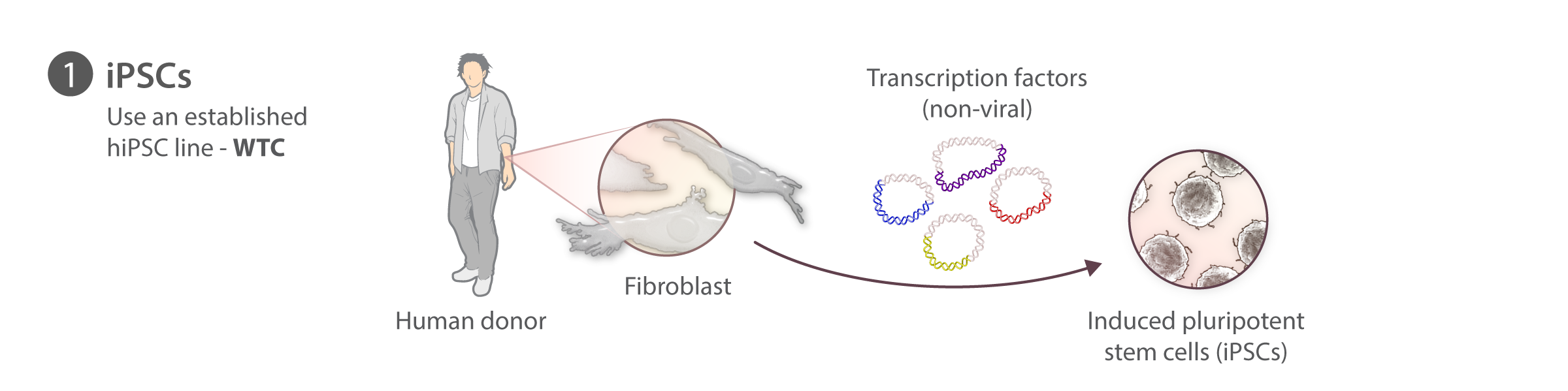
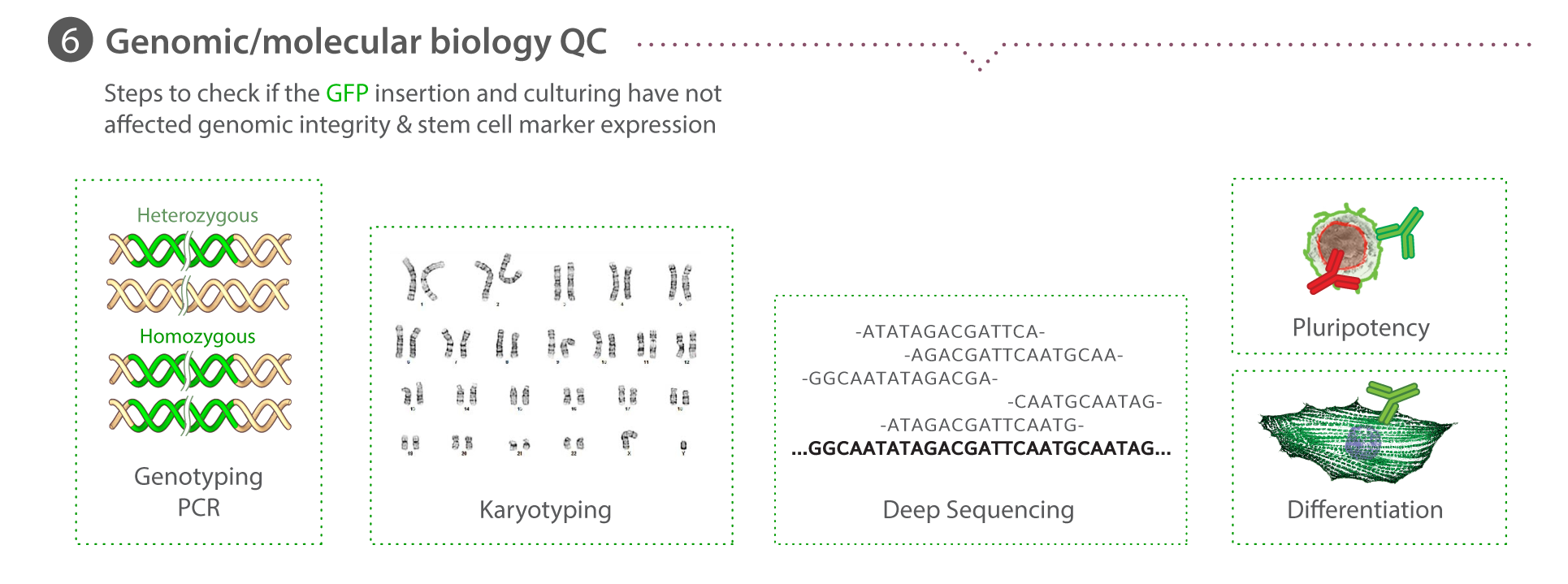
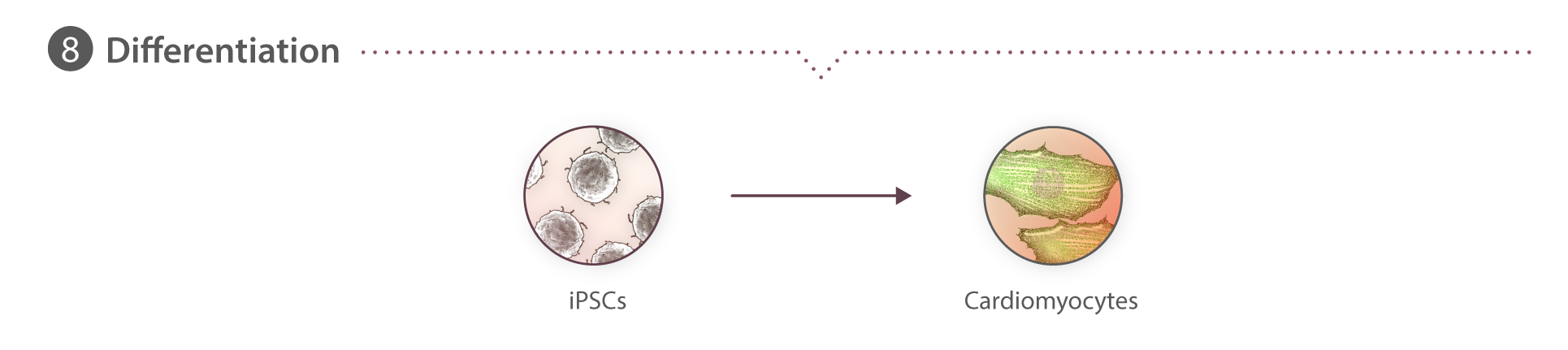
We have chosen to begin our work with hiPS cells because they are diploid, propagate in vitro, and can be induced to become nearly any kind of specialized cell, e.g., heart, kidney, nerve, skin, etc. This unique combination allows us to study multiple cell types as well as the transition from the undifferentiated to the different specialized cell types. Working with these cells also allows us to study cells derived from people with different genetic backgrounds. We chose human cells to exploit the enormous genetic information on humans that is now emerging, particularly for disease, in the wake of the Human Genome Project.
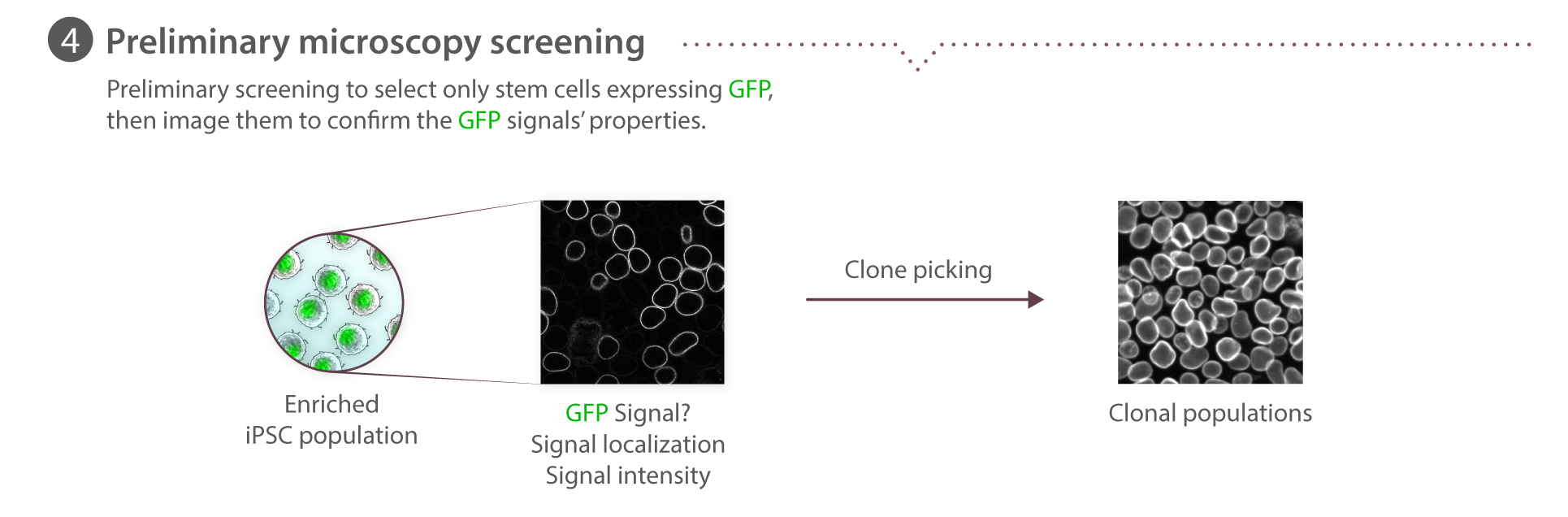
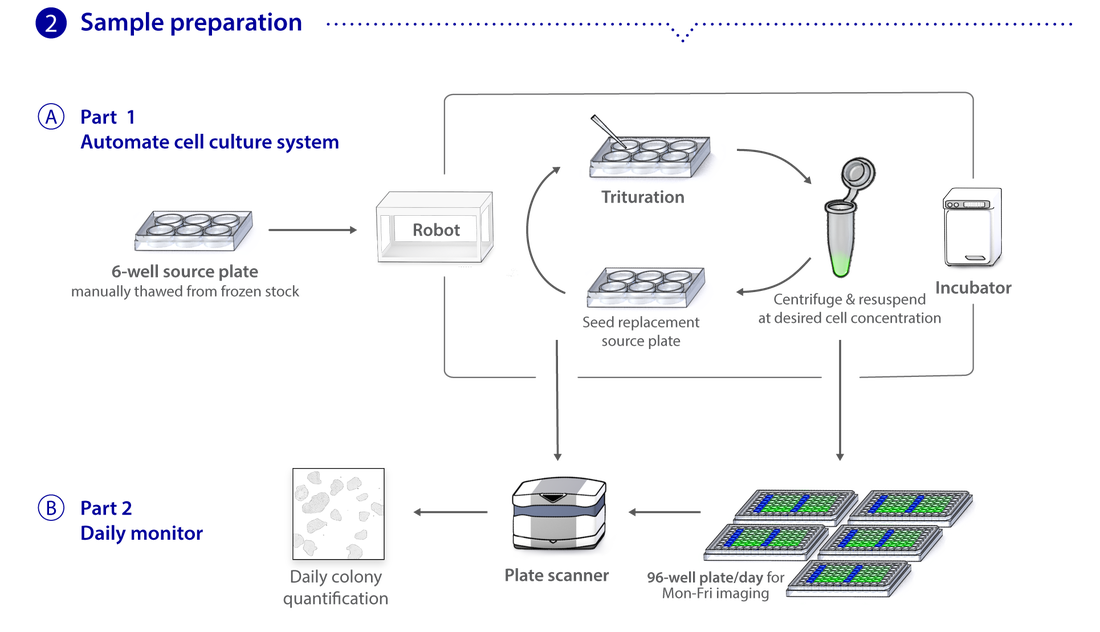
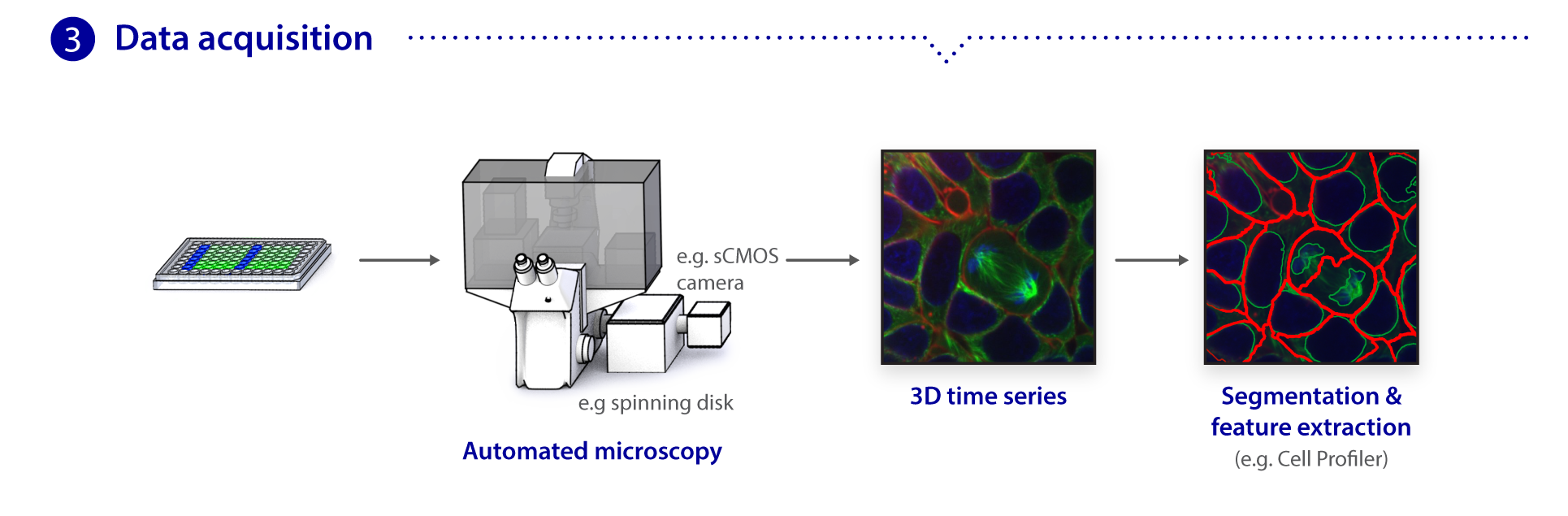
Microscopy pipeline
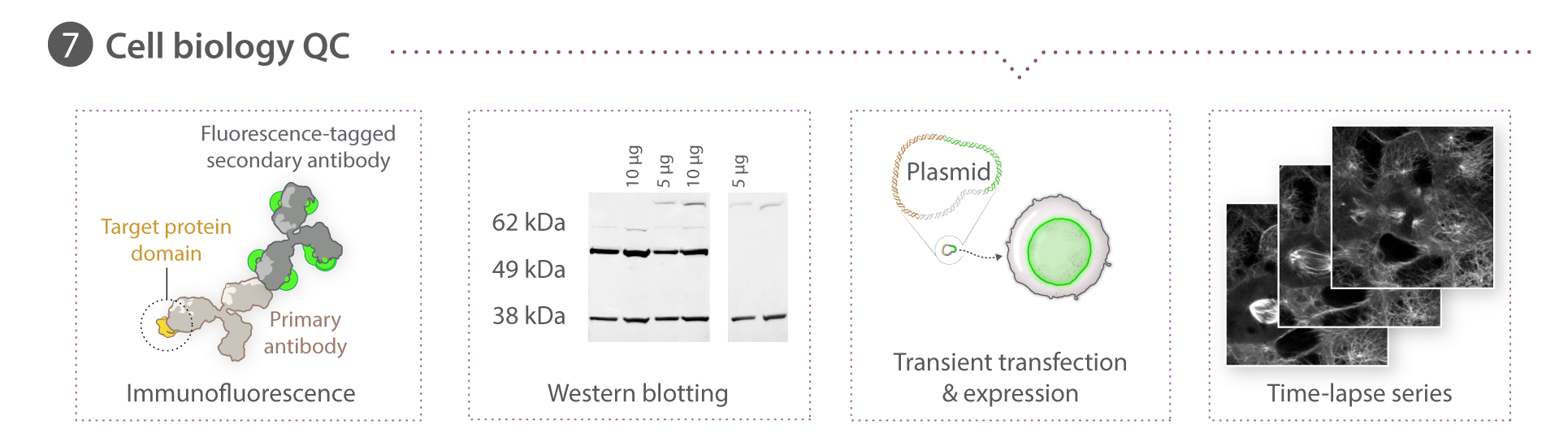
We use spinning disk and other fluorescence microscope modalities to quantify the locations and morphologies of the labeled structures in the gene-edited hiPS cells produced by Gene Editing. We automate the sample preparation and data acquisition to create large, homogeneous (replicate) data sets. The images are processed to identify and highlight (segment) the structure and features of interest, as these are most useful for modeling and statistical analyses. The data storage and tracking system is based on custom modified, open source components. Our Assay Development team develops the quantitative assays and image analysis methods; they also assess the images for biologically interesting information. The Microscopy team develops the imaging modalities and automation and also executes the high replicate imaging.
The cell images we are collecting are three dimensional, making microscopy, image analysis, and visualization more challenging. The Animated Cell team is developing rich visualization methods through which the cells can be visualized and interrogated.
The segmented images and other data are then used for computational modeling. This effort is largely statistical and correlative at present, but will also employ integrative and multi-scale approaches. Our main goals are to develop machine learning and other statistical approaches that capture the enormous cell variation that we are seeing. We are also using these methods to analyze features measured in different experiments to produce a model that integrates them into an accurate representation of the cell interior at the organelle level. This representation will reveal the organization of different cell types and states; it will also serve as a physical platform for whole-cell modeling.
All of our tools, methods, data, analyses and models are available to the public here at the Allen Cell Explorer. Our data storage and tracking system is based on custom modified, open source components.
The segmented images and other data are then used for computational modeling. This effort is largely statistical and correlative at present, but will also employ integrative and multi-scale approaches. Our main goals are to develop machine learning and other statistical approaches that capture the enormous cell variation that we are seeing. We are also using these methods to analyze features measured in different experiments to produce a model that integrates them into an accurate representation of the cell interior at the organelle level. This representation will reveal the organization of different cell types and states; it will also serve as a physical platform for whole-cell modeling.
All of our tools, methods, data, analyses and models are available to the public here at the Allen Cell Explorer. Our data storage and tracking system is based on custom modified, open source components.