|
The Allen Cell & Structure Segmenter (Segmenter) is a Python-based open source toolkit developed at the Allen Institute for Cell Science for 3D segmentation of intracellular structures in fluorescence microscope images. This toolkit brings together classic image segmentation and iterative deep learning workflows first to generate initial high-quality 3D intracellular structure segmentations and then to easily curate these results to generate the ground truths for building robust and accurate deep learning models.
The toolkit takes advantage of the high replicate 3D live cell image data collected at the Allen Institute for Cell Science of over 30 endogenous fluorescently tagged human induced pluripotent stem cell (hiPSC) lines. Each cell line represents a different intracellular structure with one or more distinct localization patterns within undifferentiated hiPS cells and hiPSC-derived cardiomyocytes. |
Getting started with Segmenter
|
Graphic interface for Segmenter
Learn how to use Segmenter via a GUI plugin for napari (details below) Jupyter notebook Look-up table demo ↗︎ A collection of workflow as a starting point |
Segmenter code on GitHub
Classic code ↗ Classic image segmentation workflow ML code ↗ Iterative deep learning workflow Model zoo ↗ Batch processing script and functions to run deep learning based segmentation models |
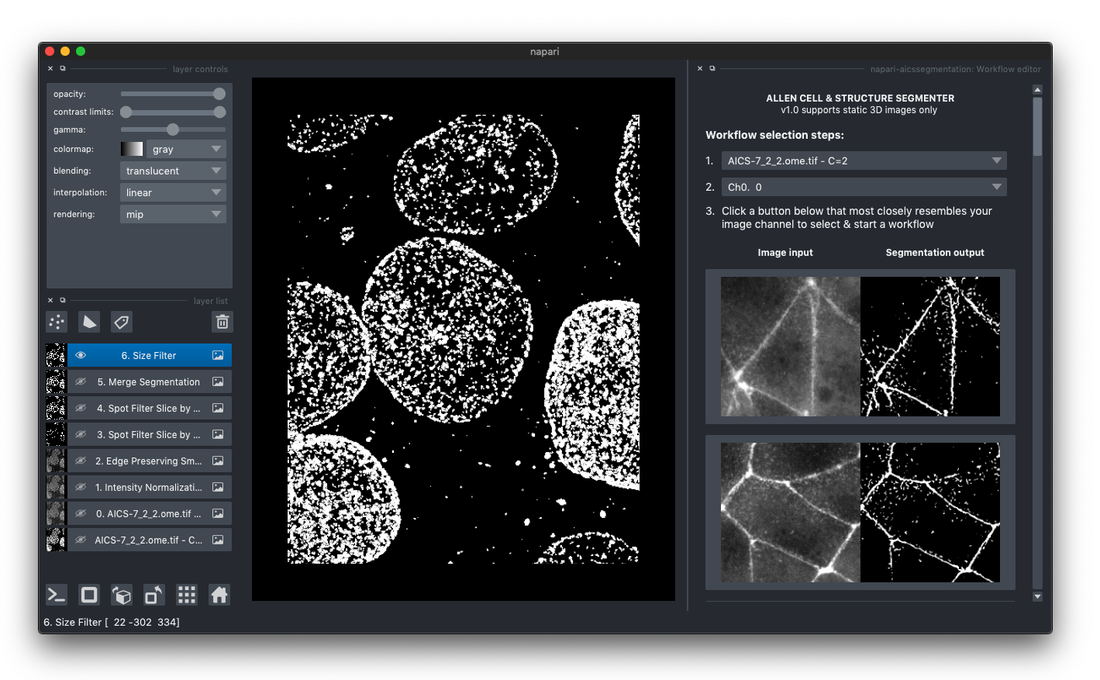
Get the Segmenter plugin
Access the 3D segmentation power of Segmenter via an intuitive graphical user interface, newly available as the Allen Cell & Structure Segmenter plugin (Segmenter plugin) for the popular open source image analysis software napari.
Version 1.0.0 (June 30, 2021)
Version 1.0.0 (June 30, 2021)
- All of the classic segmentation workflows in the look-up table are available in the plugin
- Adjust workflow parameters to maximize segmentation quality for your data
- Save/load customized workflows
- Run batch processing on default or customized workflows
Instruction
|
1. Install napari
If you don't have napari, follow napari's tutorial that walks you through various ways you can install the application. We recommend installing as a bundle app if you are a non-developer. |
2. Install the Segmenter plugin
Within the napari application, you can search for "napari-allencell-segmenter" plugin and install by accessing the menu item "Plugins" -> "Install/Uninstall Package(s)". View this video tutorial to install the plugin. |
Development notes
We continuously strive to make Segmenter more accessible to biologists and participated in the Napari Plugin Alfa Cohort to develop robust access to Segmenter’s core infrastructure and its classic segmentation workflows. The first release provides a user interface for all of the classic Segmenter workflows. In the future we plan to make the full functionality of Segmenter available in the plugin.
We continuously strive to make Segmenter more accessible to biologists and participated in the Napari Plugin Alfa Cohort to develop robust access to Segmenter’s core infrastructure and its classic segmentation workflows. The first release provides a user interface for all of the classic Segmenter workflows. In the future we plan to make the full functionality of Segmenter available in the plugin.
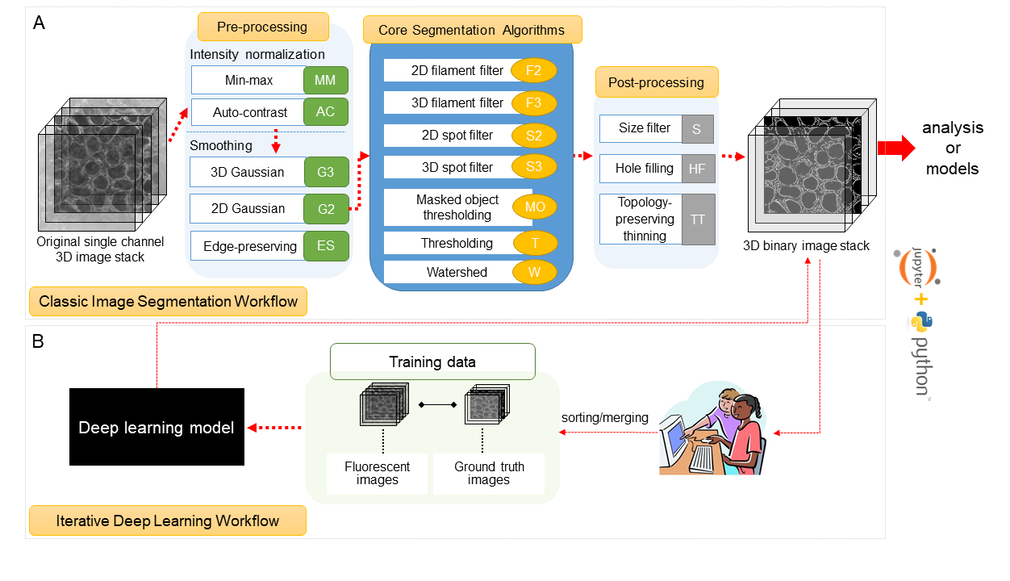
Segmenter overview
Segmenter consists of two complementary elements, a classic image segmentation workflow, with a restricted set of algorithms and parameters, and an iterative deep learning segmentation workflow. We created a collection of 20 classic image segmentation workflows based on 20 distinct and representative intracellular structure localization patterns as a “lookup table” reference and starting point for users. The iterative deep learning workflow can take over when the classic segmentation workflow is insufficient. Two straightforward “human-in-the-loop” curation strategies convert a set of classic image segmentation workflow results into a set of 3D ground truth images for iterative model training without the need for manual painting in 3D. Segmenter thus leverages state of the art computer vision algorithms in an accessible way to facilitate their application by the experimental biology researcher.
To access the toolkit, please check out our Github repositories for the Machine Learning Code & the Classic Code, as well as the accompanying paper on bioRxiv.
Note :
To access the toolkit, please check out our Github repositories for the Machine Learning Code & the Classic Code, as well as the accompanying paper on bioRxiv.
Note :
- Due to the lag between development and production cycles, the segmentation results on Data Download page are continuously being updated using our new toolkit. In the meantime, applying our ML code or Classic code on Github can produce our most recent segmentation results.
- We appreciate your feedback. Please visit the Segmenter section on image.sc discussion forum for community questions and comments.
Video tutorials
To assist biologists and computational scientists with using Segmenter, take a look at our two video tutorials:
|
Allen Cell Structure Segmenter: Creating a new classic image segmentation workflow
Our Scientific Data Engineer, Ruian Yang, walks users through the steps to create a new classic segmentation workflow for 3D image processing. |
Allen Cell Structure Segmenter: Using Iterative Deep Learning
Our Scientist, Melissa Hendershott describes the steps to combine the classic segmentation steps with deep learning, and provides helpful tips throughout the demonstration. |
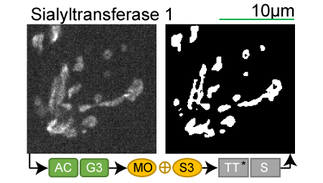
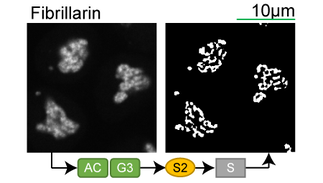
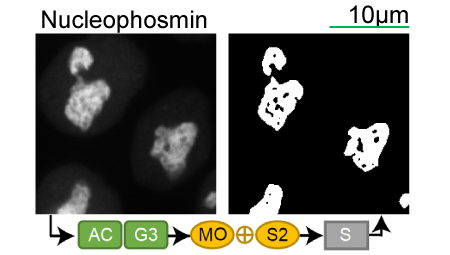
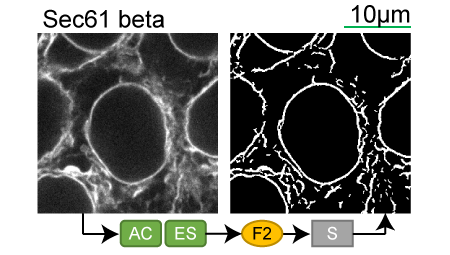
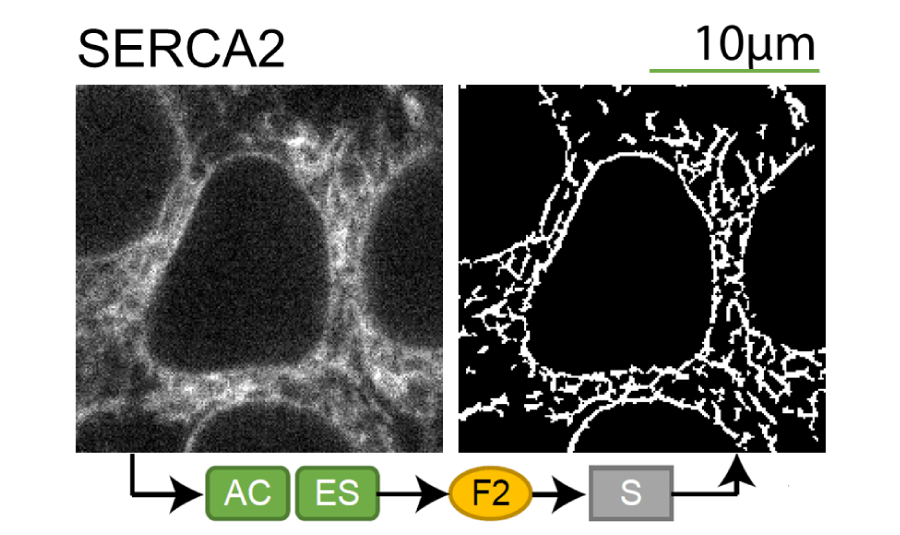
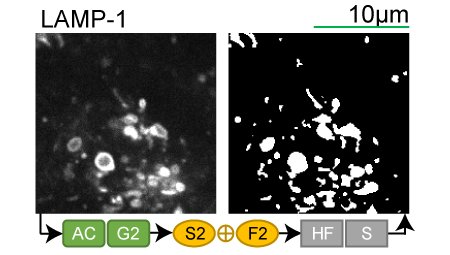
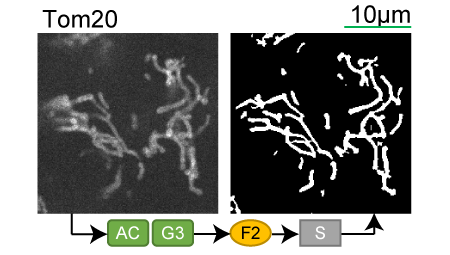
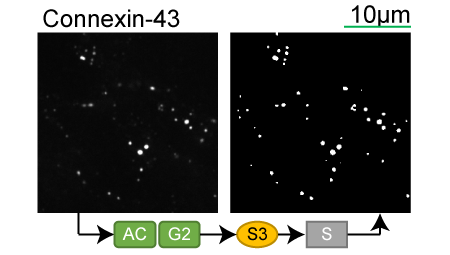
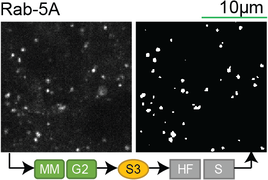
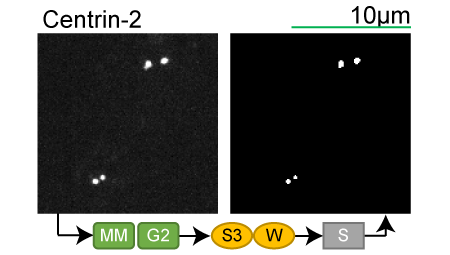
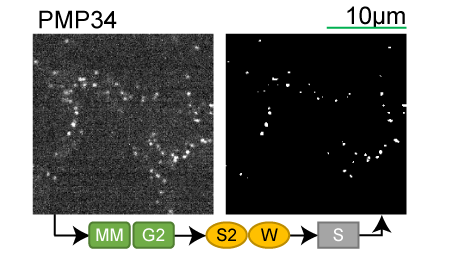
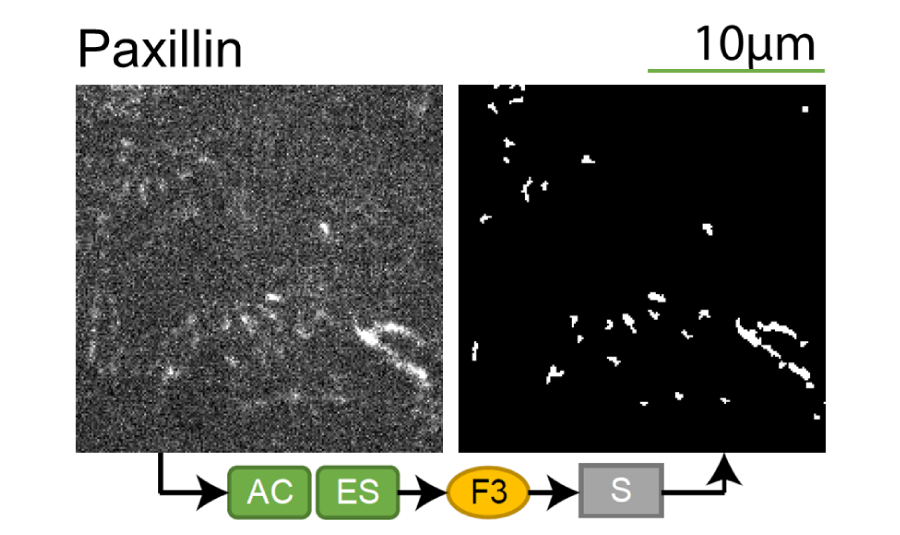
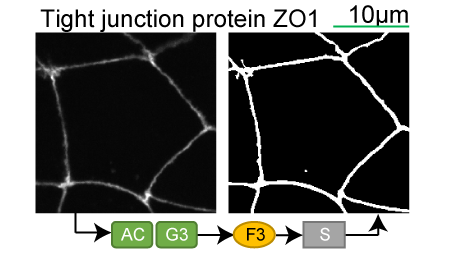
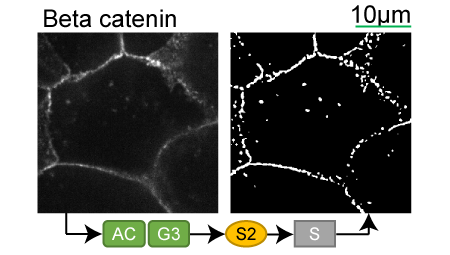
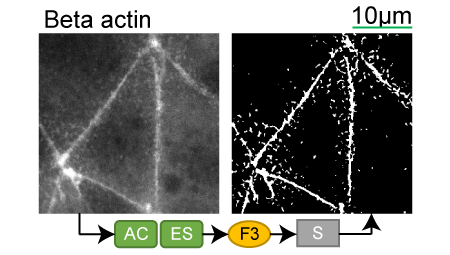
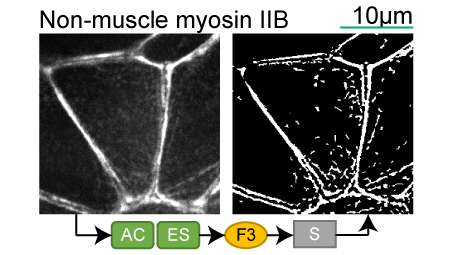
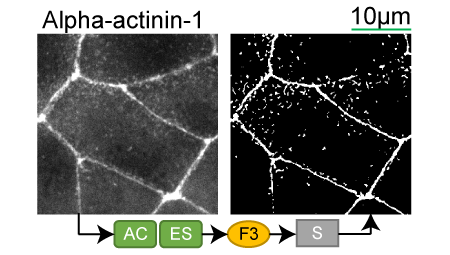
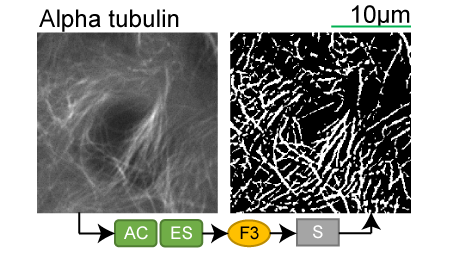
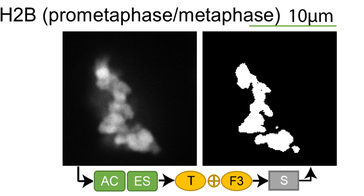
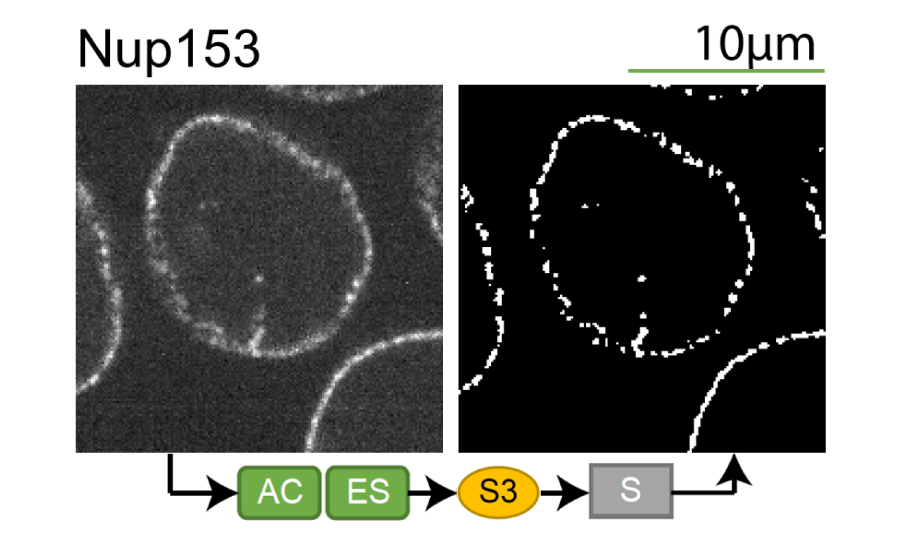
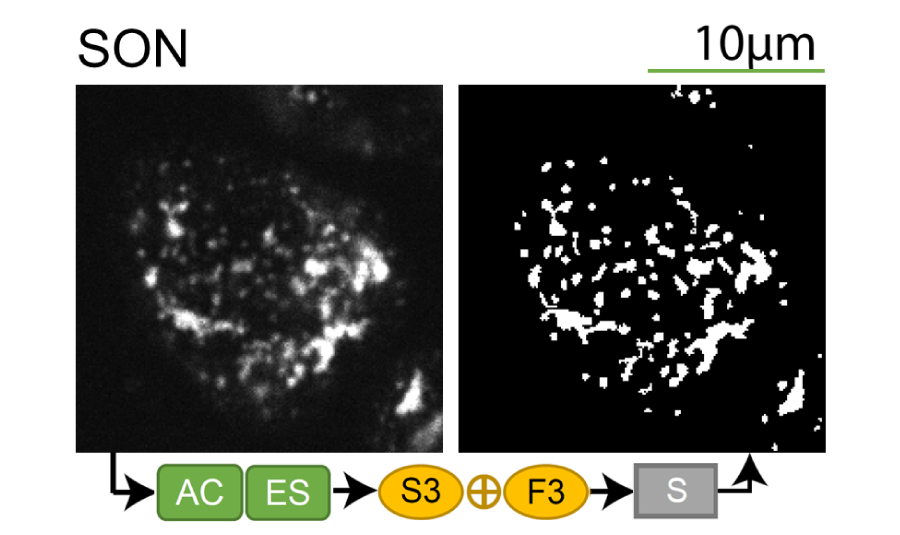
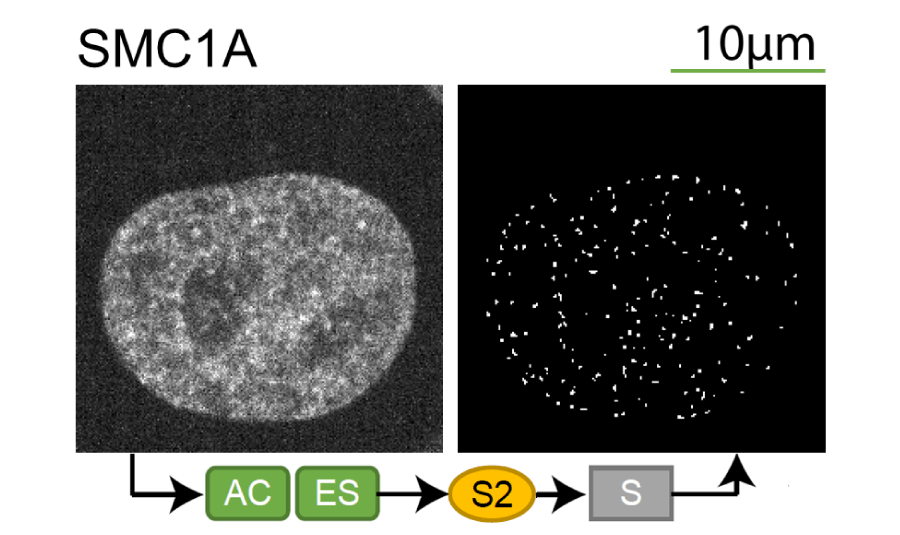
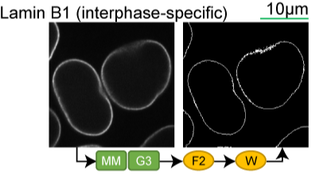
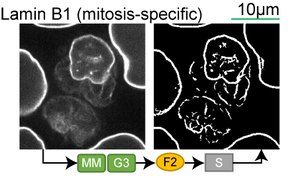
Lookup table of classic image segmentation worksflows for 28 intracellular structure localization patterns
Click on thumbnails to view
Full z-stack examples
*Note: all scale bars in these movies are 10μm
LAMP-1 |
TOM20 |
Desmoplakin |
PMP34 |
Connexin-43 |
Rab-5A |
Beta catenin |
Tight junction protein ZO1 |
Beta actin |
Non-muscle myosin IIB |
Alpha-actinin-1 |
Alpha tubulin |
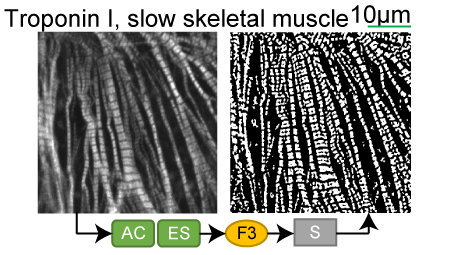
Troponin I, slow skeletal muscle |
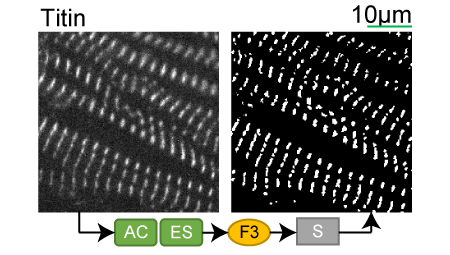
Titin |
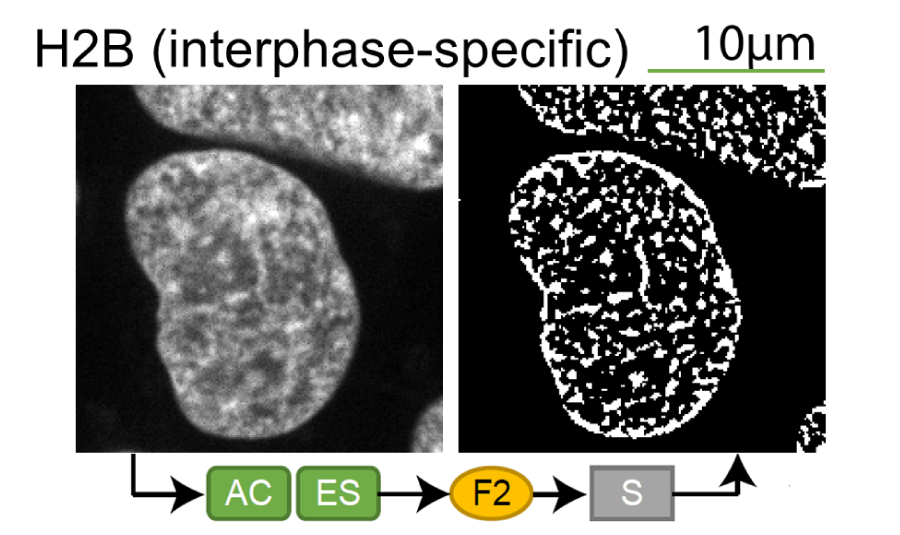
Lamin B1 (interphase-specific) |
The Institute |
Legal |
Help & contact |
Follow Us
|
Copyright © 2024 Allen Institute. All Rights Reserved.
|
|
See more on alleninstitute.org
|