What are Integrated Cell Models?
A major goal of our research is to understand 3D cell organization and how it changes over time in living cells. To do this, we use fluorescence microscopy to obtain 3D images of the major structures in living cells and then analyze the data to quantify their shapes, relative locations, amounts, etc. The great advantage of fluorescence microscopy is that it provides high quality 3D images. However, the approach only allows us to visualize a small number of cellular structures at one time due to damage caused by too much laser light. In a typical measurement, for example, we image the DNA, the cell membrane and one endogenously tagged protein designed to reveal the location and morphology of a single cellular structure. To date, we have imaged over 30 tagged structures in this way.
The challenge is how to use these data to build an integrated, holistic view of the cell. We have developed a variety of methodologies to integrate many images that each contain three or fewer structures into collections of 3D representations of single integrated cell models that each contain many (over ten) structures. The approaches include using artificial intelligence to extract information from brightfield images; transposing images of cells onto one another, either directly, statistically, or via state-of-the-art deep learning techniques; and fitting unique cells and their components into a common cell shape via spatial morphing.
The challenge is how to use these data to build an integrated, holistic view of the cell. We have developed a variety of methodologies to integrate many images that each contain three or fewer structures into collections of 3D representations of single integrated cell models that each contain many (over ten) structures. The approaches include using artificial intelligence to extract information from brightfield images; transposing images of cells onto one another, either directly, statistically, or via state-of-the-art deep learning techniques; and fitting unique cells and their components into a common cell shape via spatial morphing.
May 25, 2021: Our older content below describes two of the cell integration technologies and resulting models developed <2018. We will update this page in late 2021 with more comprehensive descriptions and comparisons of the various techniques presented for cell data integration and links to the various models and software used to generate them.
A unifying view of human induced pluripotent stem cells
|
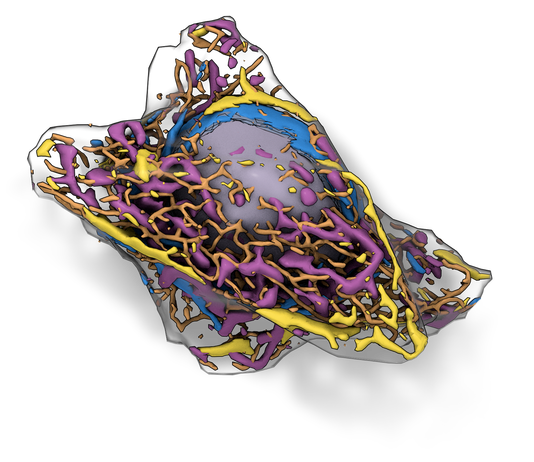
The Allen Integrated Cell is a predictive, 3D model of human induced pluripotent stem cell (hiPSC) organization. It provides a realistic, data-driven 3D visualization of a living hiPSC in its pluri-potent state. The visualization shows the many molecular machines and structures (organelles) inside the cell, simultaneously. This integrated organization drives the cell’s basic functions, and these models provide a baseline for new models of different cell types, disease, drug responses, and cellular environments.
The Allen Integrated Cell unites two technologies to improve our understanding of how human induced pluripotent stem cells (hiPSCs) vary in both shape and organization. One is a deterministic model, which shows the organization of organelles in particular individual cells, and the other model is probabilistic - showing likely shapes and locations for organelles in any cell - even for cells we have not studied. |
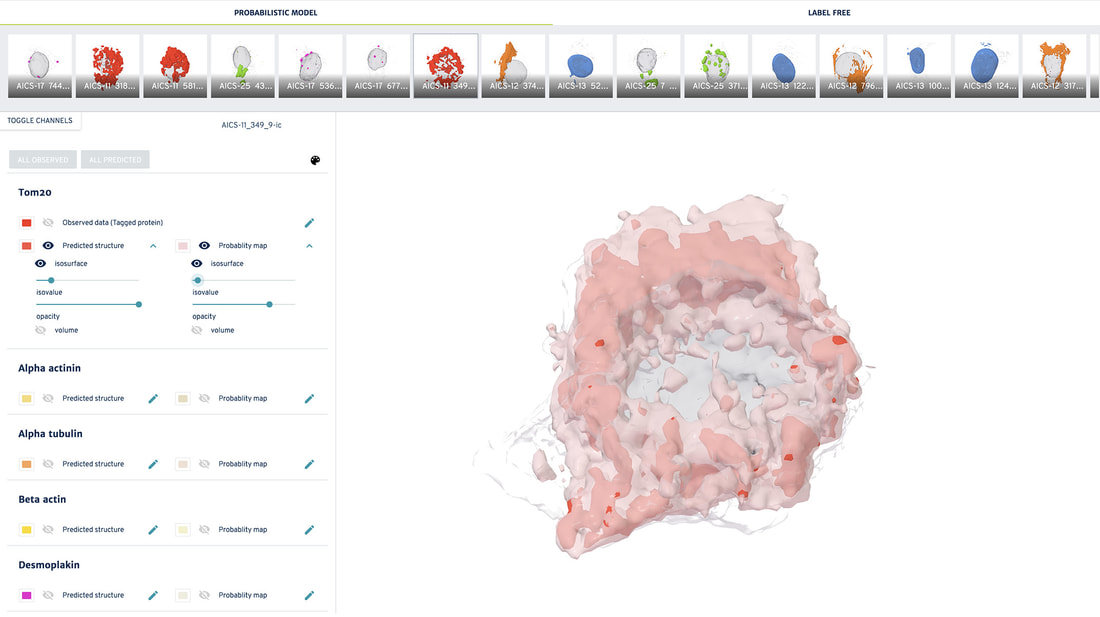
Below, you can interactively explore many examples of our cells - including some that are preparing to divide(!) - in both the 3D deterministic and probabilistic views of the Allen Integrated Cell.
How do the models work?
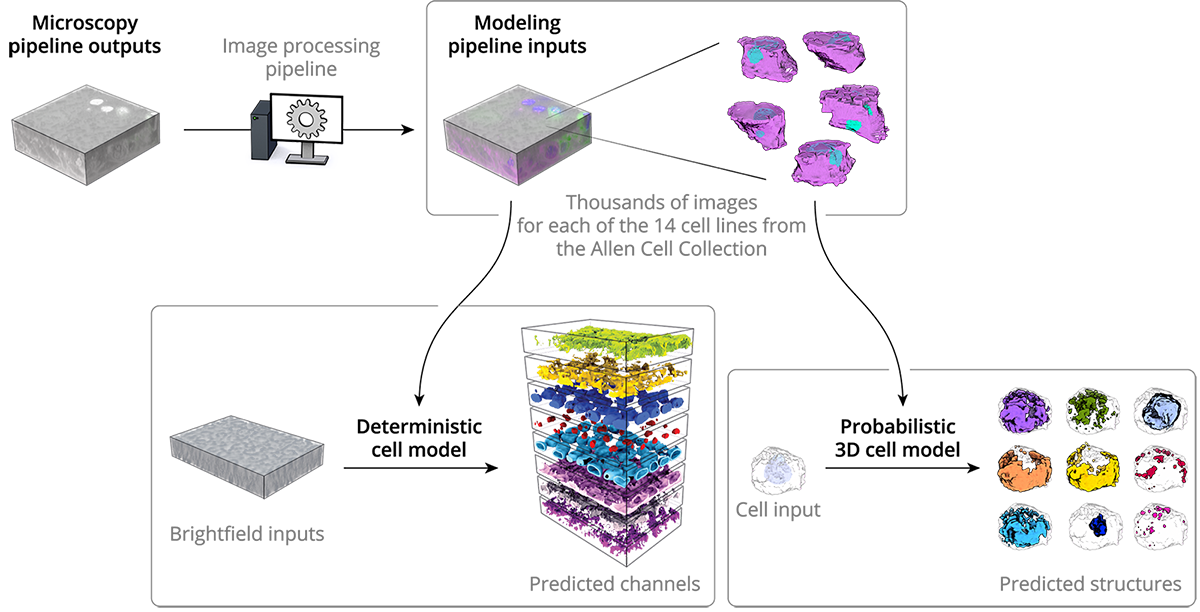
As described in our cell methods and microscopy methods pages, we collect 3D images of thousands of cells from our collection of cell lines. Each cell has one of 14 different proteins endogenously tagged with a fluorescent label, so that we can see a particular structure of interest inside of the cell. We use this data as input for both the deterministic and probabilistic models.
|
In the deterministic model, our deep learning algorithms learn the relationship between the cell structure outlined by tagged voxels and an unlabeled bright-field image, by mining these relationships from our vast database of fluorescence images for each structure. The model can then detect where an organelle is, given only a simple unlabeled brightfield image, allowing us to detect and therefore integrate many organelles from the same input image.
|
The probabilistic model, also machine learning-based, allows us to generate a probabilistic view that shows the most probable location of all of the organelles of interest for any cell, based on the location and morphologies of the cell boundary and the nucleus. This model provides an estimate of where organelles are most likely to be located in a cell and how they will look.
|
I’d really like to see the two types of models used by the Allen Integrated Cell in the same viewport...
We want you to explore deterministic and probabilistic model predictions in the same viewer, are currently implementing this, and will make this available later this year.