Observations
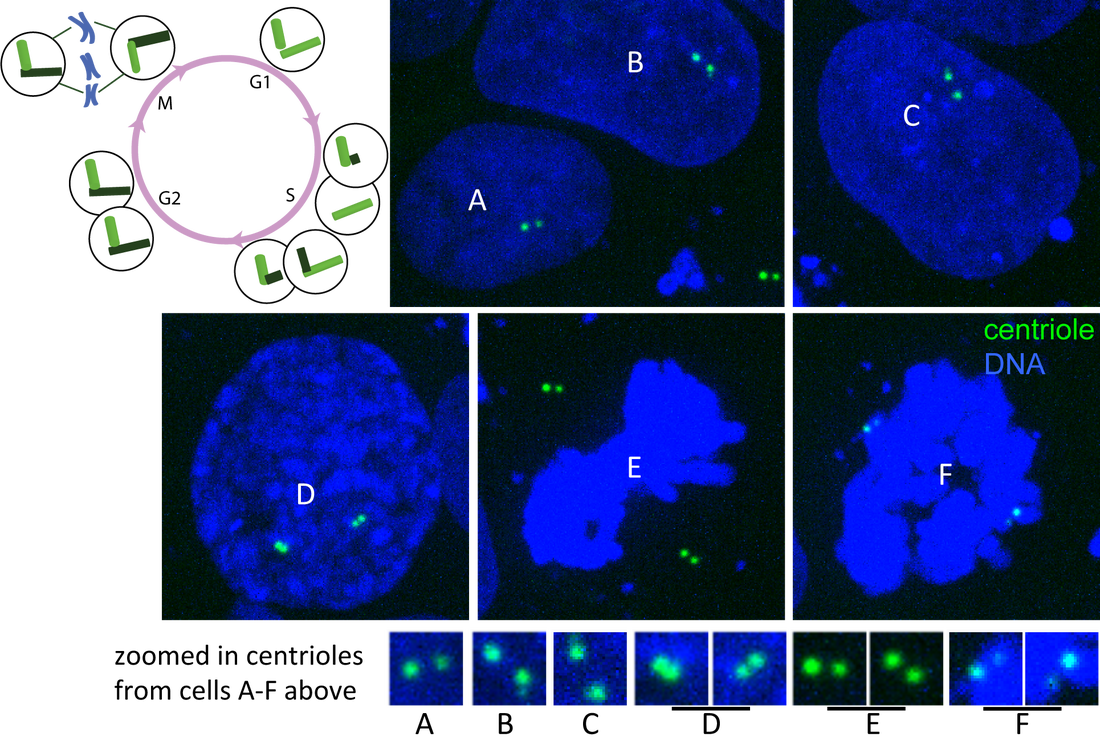
Figure. Centrin and DNA through cell cycle. Single plane images of hiPS cells expressing mTagRFP-T–tagged centrin (green) and labeled with Hoechst dye (DNA; blue) imaged on a spinning-disk confocal microscope. Cells labeled A-F represent different stages of the cell cycle (see diagram). A) G1-phase, B) early S-phase, C) later S-phase, D) G2/M-phase and E–F) M-phase contain centrioles at distinguishable stages of duplication (see zoomed in images and cell cycle diagram).
Observations
Figure. Timelapse movies of ZO-1 in tight junctions. Timelapse movies of live hiPS cells expressing mEGFP-tagged tight junction protein ZO-1 imaged on a spinning-disk confocal microscope. Images were collected in 3D every 3 min for 1.5 hrs (left) or for 15 hrs (right). Images are maximum intensity projections; playback speed is 910x (left) and 1800x (right) real time.
Observations
Z-stack with overlay Low magnification timelapse Figure. Movies of desmoplakin in desmosomes. Top: Z-stack of live hiPS cells expressing mEGFP-tagged desmoplakin imaged on a spinning-disk confocal microscope. Images start from the bottom of the cells and end at the top. The right panel shows the left panel overlaid onto the equivalent transmitted light image. Bottom: timelapse movie of a hiPS cell colony expressing mEGFP-tagged desmoplakin. Images were collected in 3D every 4 minutes for 8 hours on a spinning-disk confocal microscope. Images are maximum intensity projections; playback speed is 2400x real time.
Observations
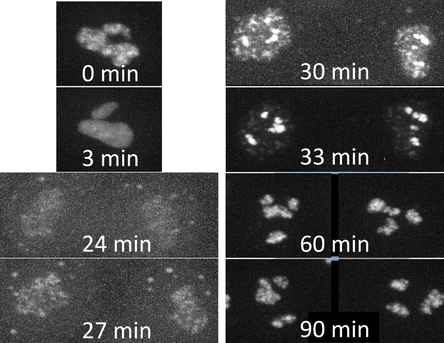
Figure 1. Movies of fibrillarin in Nucleoli. Left: Z-stack of live hiPS cells expressing mEGFP tagged fibrillarin imaged on a spinning-disk confocal microscope. Images start from the bottom of the cells and end at the top. Right: Timelapse movie of live hiPS cells expressing mEGFP tagged fibrillarin. Images were collected in 3D every 3 minutes for 1.5 hours on a spinning-disk confocal microscope. Image is a maximum intensity projection. Playback speed is 900x real time. Figure 2. Time series of cell division. A single cell going through cell division taken from the movie on the right. Observations
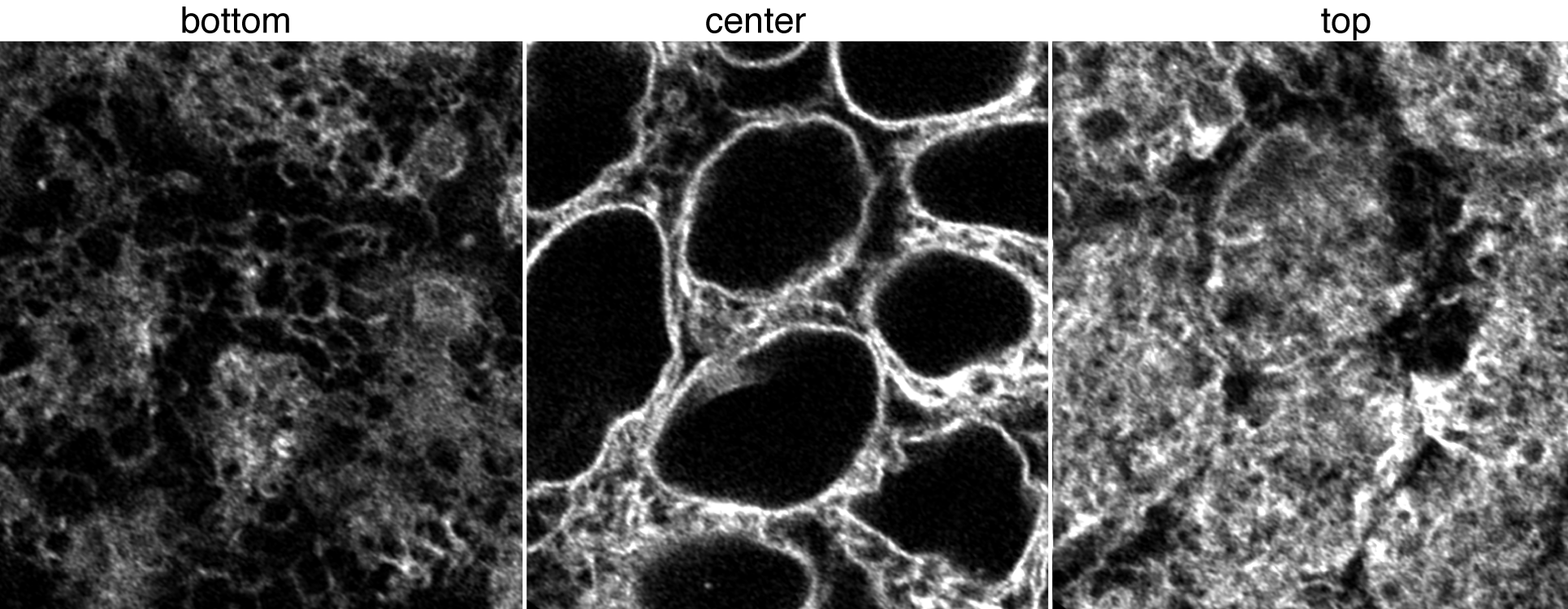
Figure 1. Movies of Sec61-ß in ER. Top left: Z-stack of live hiPS cells expressing mEGFP tagged Sec61-ß imaged on a spinning-disk confocal microscope. Images start from the bottom of the cells and end at the top. Top right: Timelapse movie of live hiPS cells expressing mEGFP tagged Sec61-ß. Images were collected in 3D every 2 minutes for 3 hours on a spinning-disk confocal microscope. Image is a single slice through the center of the cells. Playback speed is 600x real time. Bottom image panel: live hiPSC cells expressing mEGFP tagged Sec61-ß imaged on a Zeiss LSM 880 AiryScan FAST in super-resolution mode. Figure 2. Images of Sec61-ß in ER. Left, middle, and right images represent a single slice at the bottom, center, and top of cells with AiryScanFast SuperRes Observations
Figure. Live cell movies of Tom20 in Mitochondria. Left: Z-stack of live hiPS cells expressing mEGFP-tagged Tom20 imaged on a spinning-disk confocal microscope. Images start from the bottom of the cells and end at the top. Right: Timelapse movie of hiPS cells expressing mEGFP-tagged Tom20 imaged on a Zeiss LSM880 Airyscan FAST in super-resolution mode. Images were collected in 3D every 30 seconds for 30 minutes. Images show a single slice near the bottom of the cell; playback speed is 150x real time.
Observations
Figure. Movies of α-tubulin in microtubules. Top left: Z-stack of live hiPS cells expressing mEGFP-tagged α-tubulin imaged on a spinning-disk confocal microscope. Images start from the bottom of the cells and end at the top. Top center and right: Timelapse movies of a hiPSC colony expressing mEGFP-tagged α-tubulin imaged on a spinning disk confocal microscope. Center: images were collected in 3D every 4 minutes for 400 minutes. Images are maximum intensity projections; playback speed is 1200x real time. Top right: images were collected as a single slice near the top of the cell every 1 minute for 65 minutes; playback speed is 900x real time. Bottom row: 3D reconstructions of hiPS cells expressing mEGFP-tagged α-tubulin to visualize both the general organization of microtubules within the cell and the primary cilia at the top of cells.
Observations:
Figure. Timelapse movies of hiPSC cells expressing mEGFP tagged Lamin B1. Images were collected in 3D every 3 minutes for 12 hours (left) or every 35 seconds for 23 minutes (right) on a spinning-disk confocal microscope. Images are maximum intensity projection (left) or single slices from the middle of the z-stack (right). Playback speed is 1800x (left) and 350x (right) real time.
Observations
|
AboutObservations and descriptions from the microscope Archives
February 2019
Categories
All
|
The Institute |
Legal |
Help & contact |
Follow Us
|
Copyright © 2024 Allen Institute. All Rights Reserved.
|
|
See more on alleninstitute.org
|

 RSS Feed
RSS Feed