|
Movie. Z-stack and timelapse movie of nucleoli. The left panel shows a Z-stack of a live hiPS cell colony expressing mEGFP-tagged nucleophosmin. Cells were imaged in 3D on a spinning-disk confocal microscope. Right panel is the same image as the left but with contrast enhanced to visualize dimmer localization in mitotic cells. Movie starts at the bottom of the cells and ends at the top. Scale bar = 5µm. Movie. Z-stack and timelapse movie of nucleoli. Timelapse movie of a live hiPS cell colony expressing mEGFP-tagged nucleophosmin. Cells were imaged on a spinning-disk confocal microscope every 3 min. Image is a maximum intensity projection through the volume of the cells. Frames were histogram matched to adjust for photobleaching. Movie plays at 900x real time. Scale bar = 5 µm.
Observations
Figure 1. Timelapse movie of plasma membrane. Timelapse movie of a live hiPS cell colony expressing the CAAX domain of K-Ras tagged with mTagRFP-T. Cells were imaged in 3D on a spinning-disk confocal microscope every 5 min. Panels show the same colony at different z-positions. Left) The bottom z-section of the cells. Middle) A maximum intensity projection through the middle 5 z-sections of the cells. Right) A maximum intensity projection through the top 12 z-sections of the cells. Frames in each panel were histogram matched to adjust for photobleaching. Movie plays at 1500x real time. Scale bar = 5 µm. Figure 2. Timelapse movie of plasma membrane. Timelapse movie of a live hiPS cells in an epithelial sheet containing both unedited cells (which remain unseen in black), and cells expressing the CAAX domain of K-Ras tagged with mTagRFP-T. Cells were imaged in 3D on a spinning-disk confocal microscope every 3 min; the top and bottom z-sections are shown pseudocolored in green and magenta, respectively. Movie plays at 1800x real time. Scale bar = 10 µm.
Observations
Figures. Z-stack and timelapse movies of endosomes. A) Z-stack of a live hiPS cell colony expressing two copies (biallelic) of mEGFP-tagged Ras-related protein Rab-5A imaged in 3D on a spinning-disk confocal microscope. Movie starts at the bottom of the cells and ends at the top. Scale bar = 5µm. B) Timelapse movie of a live hiPS cell colony expressing two copies (biallelic) of mEGFP-tagged Ras-related protein Rab-5A. Cells were imaged on a spinning-disk confocal microscope. A single, mid-level plane was acquired every second. A zoom-in of the area boxed in the left panel is shown on the right. Movie sped up 10x real time. Scale bar = 5 µm.
Observations
Figures. Z-stack and timelapse movies of peroxisomes. A) Z-stack of a live hiPS cell colony expressing mEGFP-tagged peroxisomal membrane protein PMP34 imaged in 3D on a spinning-disk confocal microscope. Movie starts at the bottom of the cells and ends at the top. Scale bar = 5 µm. B) Timelapse movie of a live hiPS cell colony expressing mEGFP-tagged peroxisomal membrane protein PMP34. Cells were imaged on a spinning-disk confocal microscope every 1s. Movie sped up 10x real time. Images are shown with the despeckle ImageJ filter applied. Scale bar = 5 µm.
Observations
Figures. Z-stack and timelapse movies of gap junctions. A) Z-stack of a live hiPS cell colony expressing mEGFP-tagged connexin-43. Cells were imaged in 3D on a spinning-disk confocal microscope. Movie starts at the bottom of the cells and ends at the top. Scale bar = 5 µm. B) Timelapse movie of a live hiPS cell colony expressing mEGFP-tagged connexin-43. Cells were imaged on a spinning-disk confocal microscope every 3.25 min. Image is a maximum intensity projection of 20 z-slices acquired at the top of the cells. Contrast is enhanced to show localization of connexin-43 to internal structures. Frames were histogram matched to adjust for photobleaching. Movie plays at 975x real time. Scale bar = 5 µm.
Observations
Golgi visualized via sialyltransferase 1 (also known as ß-galactoside α-2,6-sialyltransferase1)10/5/2017
Figure 1. Z-stack movies of Golgi. A) Z-stack of live hiPS cells expressing one copy (monoallelic) of mEGFP-tagged sialyltransferase 1 (ST6GAL1) imaged on a spinning-disk confocal microscope. Movie starts at the bottom of the cells and ends at the top. B) Z-stack of live hiPS cells expressing two copies (biallelic) of mEGFP-tagged sialyltransferase 1 (ST6GAL1) imaged on a spinning-disk confocal microscope. Movie starts at the bottom of the cells and ends at the top. Contrast settings for these two movies are identical to highlight the increased signal in the biallelic edited line (right). C) Z-stack of live hiPS cells expressing two copies (biallelic) of mEGFP-tagged sialyltransferase 1 (ST6GAL1; white) stained for DNA (NucBlue Live; cyan) and plasma membrane (CellMask Deep Red; magenta). Movie starts at the bottom of the cells and ends at the top.
Figure 2. Timelapse movies of Golgi. A) Timelapse movie of hiPS cells expressing two copies (biallelic) of mEGFP-tagged sialyltransferase 1 (ST6GAL1). Images were taken in 3D every 3 min for 6 hrs on a spinning-disk confocal microscope. Images are maximum intensity projections with the ‘despeckle’ ImageJ filter applied. Playback speed is 1800x real time. B) Timelapse movie of hiPS cells expressing two copies (biallelic) of mEGFP-tagged sialyltransferase 1 (ST6GAL1). Images were taken in 3D every 5 min for 100 min on a spinning-disk confocal microscope. Images are maximum intensity projections with the ‘despeckle’ ImageJ filter applied. Playback speed is 900x real time. Observations
Figures. Z-stack and timelapse movies of lysosomes. A) Z-stack of live hiPS cells expressing mEGFP tagged LAMP1 imaged on a spinning-disk confocal microscope. Movie starts at the bottom of the cells and ends at the top. Scale bar = 5µm. B–C) Timelapse movie of live hiPS cells expressing mEGFP-tagged LAMP1. Images were taken as a single slice near the top (B) or bottom (C) of the cell every 1 sec for 100 sec on a spinning-disk confocal microscope. Images have had the ‘despeckle’ ImageJ filter applied. Movie sped up 10x real time. Scale bar = 5 µm.
Observations
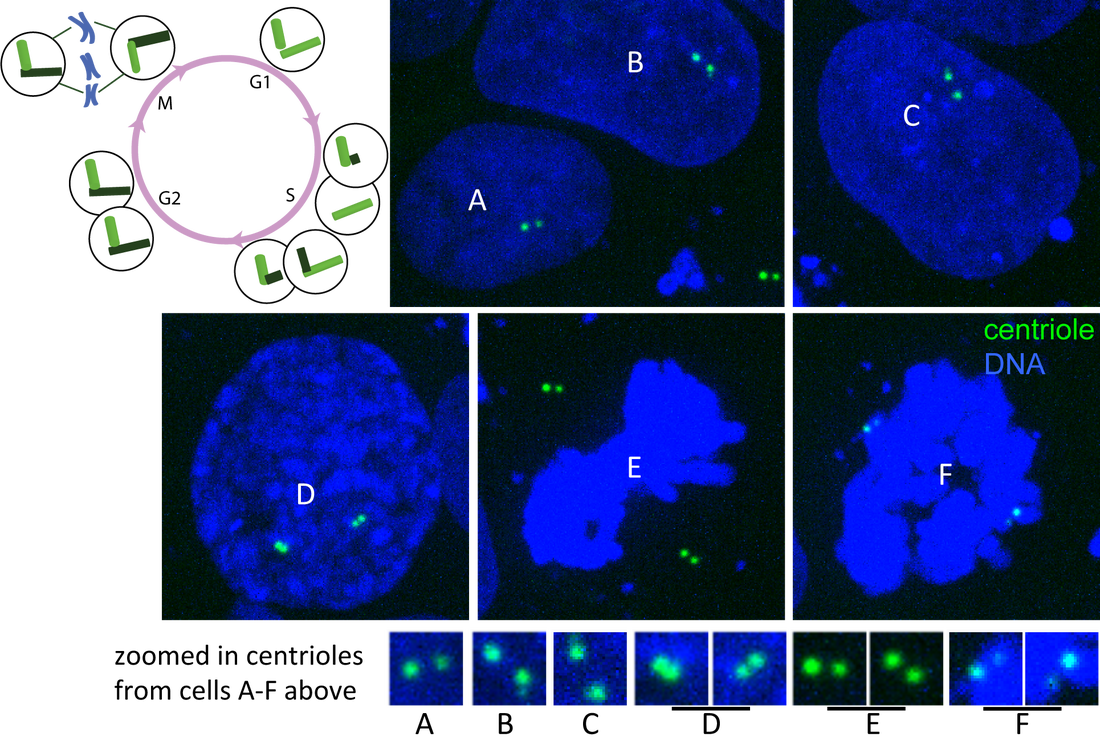
Figure. Centrin and DNA through cell cycle. Single plane images of hiPS cells expressing mTagRFP-T–tagged centrin (green) and labeled with Hoechst dye (DNA; blue) imaged on a spinning-disk confocal microscope. Cells labeled A-F represent different stages of the cell cycle (see diagram). A) G1-phase, B) early S-phase, C) later S-phase, D) G2/M-phase and E–F) M-phase contain centrioles at distinguishable stages of duplication (see zoomed in images and cell cycle diagram).
Observations
Figure. Timelapse movies of ZO-1 in tight junctions. Timelapse movies of live hiPS cells expressing mEGFP-tagged tight junction protein ZO-1 imaged on a spinning-disk confocal microscope. Images were collected in 3D every 3 min for 1.5 hrs (left) or for 15 hrs (right). Images are maximum intensity projections; playback speed is 910x (left) and 1800x (right) real time.
Observations
Z-stack with overlay Low magnification timelapse Figure. Movies of desmoplakin in desmosomes. Top: Z-stack of live hiPS cells expressing mEGFP-tagged desmoplakin imaged on a spinning-disk confocal microscope. Images start from the bottom of the cells and end at the top. The right panel shows the left panel overlaid onto the equivalent transmitted light image. Bottom: timelapse movie of a hiPS cell colony expressing mEGFP-tagged desmoplakin. Images were collected in 3D every 4 minutes for 8 hours on a spinning-disk confocal microscope. Images are maximum intensity projections; playback speed is 2400x real time.
Observations
|
AboutObservations and descriptions from the microscope Archives
February 2019
Categories
All
|
The Institute |
Legal |
Help & contact |
Follow Us
|
Copyright © 2024 Allen Institute. All Rights Reserved.
|
|
See more on alleninstitute.org
|

 RSS Feed
RSS Feed